A natural resource covering almost 97% of our earth,
Making up more than two-thirds of the human body weight!
As vital as this fluid is for sustenance of life,
It can become a life-threatening noxious liquid!
Water is an abundant resource, but access to safe drinking water is still a luxury not available to one and all! People depend on water bodies, such as rivers, ponds, and wells, for abstraction of water, but these water bodies are often contaminated with noxious chemicals and/or harmful disease-causing pathogens. As per the World Health Organisation, at least 2 billion people around the world use faecal-contaminated water source for drinking purposes. Diseases like diarrhoea, cholera, dysentery, typhoid and poliomyelitis caused by a number of bacterial, viral, and parasitic organisms can be transmitted by consumption of microbiologically contaminated water and leads to approximately 4,85,000 diarrhoeal deaths every year.1
Water-borne diseases are a global burden and claim millions of lives every year. In developing countries, these diseases are associated with physical scarcity of water, i.e., lack of available usable resources as well as economic scarcity of water, that is, lack of investment in water infrastructure. Water-borne infections are caused due to ingestion, airborne exposure or contact with water contaminated by a variety of agents like bacteria, viruses, protozoa and helminths. Based on the mode of transmission, water-borne diseases have been classified into four categories, explained in Figure 1.2

Caption– Figure- 1: Categories of Water-borne Diseases
There has been a sharp decline in water-borne outbreaks since the last century, but infectious water-borne diseases are still a cause for global concern. Protozoans belonging to genera, such as Cryptosporidium, Naegleria and Giardia, and bacterial genera, such as Salmonella, Vibrio, Legionella, Escherichia, Campylobacter, Shigella and Pseudomonas, are the major causative agents associated with outbreaks and infections. Various factors such as infrastructure, chemical coats on pipes, architecture of systems, treatment deficiencies as well as weather and temperature contribute to such outbreaks.1,2 Starting with water-borne pathogens and diseases, let’s understand novel approaches in the detection of water-borne diseases.
Emerging Water-borne Pathogens
There are more than 1000 species of pathogens including bacteria, viruses, protozoa as well as fungi. The development of a disease depends on various factors such as minimal infectious dose, pathogenicity, host susceptibility and environmental characteristics. Enteric bacteria have minimal infectious dose value in the range of 107-108 cells, while certain species have it as low as 101 to 103. The species belonging to genera like Shigella, Campylobacter, Escherichia and Vibrio are known to have lower values. In case of protozoans, a few oocysts are sufficient for producing the disease, and a small number of viruses are enough for developing the disease. Viruses such as caliciviruses, enteroviruses, coxsackieviruses, polyomaviruses and cytomegalovirus are classified as water-transmitted viral pathogens. Influenza virus and coronaviruses have been suggested as organisms that can be transmitted via drinking water; however, the evidence is inconclusive.2,3
Water-borne pathogens appear repeatedly due to the following reasons:2
- Contaminated water
- Increased population sensitive to the pathogens
- Altered drinking water treatment technology
- Globalization of commerce and travel
- Development of molecular methods
Certain water-borne pathogens such as protozoa Microsporidia, bacteria (like Mycobacterium avium, Helicobacter pylori, Tsukamurella and Cystoisospora belli) and viruses (like adenoviruses, parvoviruses, coronaviruses, and polyomavirus) are examples of emerging potential in water-borne pathogens. Majority of these organisms have certain resistance against common disinfectants like chlorine, ultraviolet light and heat used for treating water sources. This resistance presents a challenge in removal of these pathogens.3 Figure 2 depicts some major pathogens that are detected in drinking water systems.1,2
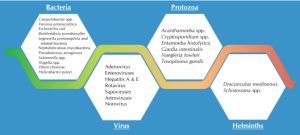
Caption– Figure- 2: Pathogens in Drinking Water Systems (Elaborated as follows:)
Bacteria
- Campylobacter
- Yersinia enterocolitica
- Escherichia coli
- Burkholderia pseudomallei
- Legionella pneumophilaand related bacteria
- Nontuberculous mycobacteria
- Pseudomonas aeruginosa
- Salmonella
- Shigella
- Vibrio cholerae
- Helicobacter pylori
Virus
- Adenovirus
- Enteroviruses
- Hepatitis A and E viruses
- Rotavirus
- Sapoviruses
- Astroviruses
- Norovirus
Protozoa
- Acanthamoeba
- Cryptosporidium
- Entamoeba histolytica
- Giardia intestinalis
- Naegleria fowleri
- Toxoplasma gondii
Helminths
- Dracunculus medinensis
- Schistosoma
Diagnosing Water-borne Pathogens
Water-borne pathogens include bacteria, viruses as well as protozoans. Reliable analysis of these requires a method that has attributes such as automation, high specificity and sensitivity, result reproducibility, as well as affordability and the time required for analysis. Staring with molecular techniques, let’s acquaint ourselves with the cutting-edge methodologies used for detection of water-borne pathogens.2
- Molecular-based Methods
These methods can be species-specific and provide phylogenetic information. Alternative indicators can also be incorporated permitting distinction between human and animal pathogens as well as tracking the source of pollution.2
- Polymerase Chain Reaction
Polymerase chain reaction (PCR) is one of the most commonly used techniques that operates by amplifying a specific target DNA sequence in a cyclic three-step process for exponential amplification of the target sequence. Quick analysis is one of the major advantages of molecular techniques. However, they require accurate primers and optimal reaction mixture for avoiding false results and cannot differentiate viable and non-viable cells. Multiplex PCR, a type of PCR allows simultaneous detection of several target organisms of diverse pathogens in a single reaction. Quantitative real-time PCR is another version of PCR that provides high sensitivity and specificity, faster rate of detection as well as minimizes the risk of cross-contamination and does not require post-PCR analysis. Quantitative reverse-transcriptase PCR provides an estimation of the concentration of RNA viruses. It can detect viable cells as it detects messenger RNA which is present only in viable organisms but might be unable to detect damaged genomes.1,2
- DNA Microarrays
Microarrays are glass slides or chips coated with specific chemically synthesized oligonucleotide probes that allow rapid detection of multiple genes of multiple organisms simultaneously owing to their capacity for screening large numbers of sequences. They have numerous advantages including high throughput capacity, automation, ability to detect antimicrobial resistance and host origin of contaminants. However, it comes with drawbacks, such as relatively high cost, low specificity and sensitivity due to non-specific hybridization and difficulty in distinguishing between viable and non-viable cells.
Oligonucleotide microarray, a powerful genomic technology, is widely used to monitor gene expression under different cell growth conditions, detect specific mutations in DNA sequences as well as characterise micro-organisms in environmental samples. DNA microarrays contain high-density immobilized nucleic acids in a two-dimensional ordered matrix that enables simultaneous detection of genes in a single assay via nucleic acid hybridization.1,2
- Next-generation Sequencing
Next-generation Sequencing (NGS) is a DNA-sequencing technology that allows high-throughput parallel analysis of DNA sequences from PCR amplicons and determines specific microbial consortia in a water sample.1
- Pyrosequencing
Pyrosequencing is a DNA sequencing technology that uses enzyme-coupled reaction and bioluminescence (polymerase, Adenosine-5 -triphosphate sulfurylase, luciferase and apyrase) to monitor pyrophosphate release in real time. This technology provides a large number of sequences read in a single run, but is limited due to cost, amount of DNA in the samples, complexity of analysis, availability of computing power and the efficiency of data generation.1,2
- Fluorescence in Situ Hybridization
Fluorescence in situ hybridization (FISH) is a technique that involves the hybridization of sample with fluorescent-labelled rRNA oligonucleotide probes which allow enumeration of particular microbial cells using confocal microscopy, fluorescence microscopy or flow cytometry. It aids in the detection and identification of different micro-organisms in mixed populations like biofilms. Major drawbacks of this technique are low sensitivity and necessity of pre-enrichment and concentration steps that might increase inhibitor concentrations leading to false-negative results.1,2
- Immunology-based Methods
Immunological detection with antibodies methods are based on antibody-antigen interaction that have been employed for the detection of multiple pathogens. Serum neutralization tests, enzyme-linked immunosorbent assays, immunofluorescence and immunomagnetic separation are some examples of immunology-based methods. The limitations associated with these methods are low sensitivity, false-negative results, cross-reactivity and need for pre-enrichment.1,2
- Biosensors
Biosensors are made of a bioreceptor (recognising target) and a transducer (converting biological interactions into measurable electrical signal). They have numerous advantages like automation, short analysis time, portability, real-time measurements, miniaturisation of biological analytical techniques and no need for pre-enrichment. The drawbacks include great sensitivity to pH, mass and temperature. Biosensors are of various types, namely:1,2
- Optical biosensors
- Electrochemical biosensors
- Mass-sensitive biosensors
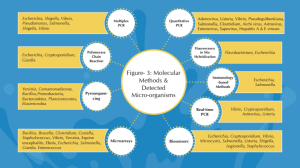
Caption– Figure- 3: Molecular Methods and Detected Micro-organisms
Hurdles in Diagnosing Water-borne Pathogens
The major challenge for the detection of water-borne pathogens is the absence or lack of a unified method encompassing water analysis for every pathogenic micro-organism of interest, such as bacterium, virus or protozoan. Some of the other major challenges in detection of water-borne pathogens include:2
- Physical differences between the major pathogen groups
- Low pathogen concentration necessitating enrichment and concentration
- Presence of inhibitors
- Absence of general protocols for sample collection
- Absence of culture-independent detection methods
- Detection of the host origin of pathogens
Quantitative Microbial Risk Assessment
Quantitative microbial risk assessment (QMRA) is a systematic quantitative assessment process that combines information about the dose response of an infectious agent with the distribution of exposure routes. QMRA aids in estimating the health risks or rates of illness due to exposure to particular pathogens. It aids in predicting the burden of water-borne diseases, setting tolerable limits, identifying means of reducing the risk and determining measures to protect water safety. There are 5 steps in risk assessments, namely:
- Hazard identification
- Exposure assessment
- Dose-response assessment
- Risk characterization
- Risk management and communication
Major drawbacks of QMRA approach are its dependency on the method used for monitoring pathogens in water and the lack of available data (from cultivation methods) for completing the assessment. QMRAspot, a computational tool based on QMRA, was developed for rapid and automatic quantitative microbial risk assessment of drinking water.2
Expanded Threat!
Water is a basic necessity, but a large population lacks access to safe clean and contamination-free water. Water-borne pathogens present a critical global health concern, especially in developing countries, making it imperative to develop different approaches for detection of pathogens. Various methods, such as cultural, molecular as well as biosensor-based, have been implemented, but they come with certain drawbacks. Culture-dependent methods are extensively used for the detection of pathogens but are time-consuming and have low sensitivity. Molecular or biosensor-based methods are dependent on lab methods but might require pre-treatments and are expensive.2
Water-borne diseases are directly related to environmental deterioration and pollution. Despite continued efforts for maintaining the safety of water, outbreaks related to water-borne pathogens are still being reported globally. As per the World Health Organisation, more than 2 billion people reside in water-stressed areas, which are expected to aggravate in the view of aggravating climate change and increasing population worldwide. With the arrival of the monsoon season, water-borne pathogens and resulting infections would be on the rise. The world is already employing precautions against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), but prevention of water-borne as well as air-borne pathogens is also of the essence!
References
- Srivastava, K. R., Awasthi, S., Mishra, P. K., & Srivastava, P. K. (2020). Biosensors/molecular tools for detection of waterborne pathogens.Waterborne Pathogens, 237-277. https://doi.org/10.1016/B978-0-12-818783-8.00013-X
- Ramírez-Castillo, F. Y., Loera-Muro, A., Jacques, M., Garneau, P., Avelar-González, F. J., Harel, J., & Guerrero-Barrera, A. L. (2015). Waterborne pathogens: detection methods and challenges.Pathogens, 4(2), 307-334. https://doi.org/3390/pathogens4020307
- Gall, A. M., Mariñas, B. J., Lu, Y., & Shisler, J. L. (2015). Waterborne viruses: a barrier to safe drinking water.PLoS pathogens, 11(6), e1004867. https://doi.org/1371/journal.ppat.1004867
Writer & Reviewer: Kayshu Grover & Dr Vaishali Khudkhudia